Pseudogene Discovery Pains Evolutionary Paradigm
It was one of the most painful experiences I ever had. A few years ago, I had two back-to-back bouts of kidney stones. I remember it as if it were yesterday. Man, did it hurt when I passed the stones! All I wanted was for the emergency room nurse to keep the Demerol coming.
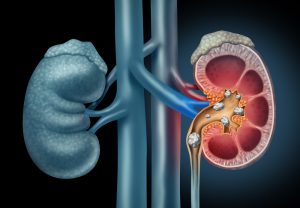
Figure 1: Schematic Depiction of Kidney Stones Moving through the Urinary Tract. Image Credit: Shutterstock
When all that misery was going down, I wished I was one of those rare individuals who doesn’t experience pain. There are some people who, due to genetic mutations, live pain-free lives. This condition is called hypoalgesia. (Of course, there is a serious downside to hypoalgesia. Pain lets us know when our body is hurt or sick. Because hypoalgesics can’t experience pain, they are prone to serious injury, etc.)
Biomedical researchers possess a keen interest in studying people with hypoalgesia. Identifying the mutations responsible for this genetic condition helps investigators understand the physiological processes that undergird the pain sensation. This insight then becomes indispensable to guiding efforts to develop new drugs and techniques to treat pain.
By studying the genetic profile of a 66-year-old woman who lived a lifetime with pain-free injuries, a research team from the UK recently discovered a novel genetic mutation that causes hypoalgesia.1 The mutation responsible for this patient’s hypoalgesia occurred in a pseudogene, a region of the genome considered nonfunctional “junk DNA.”
This discovery adds to the mounting evidence that shows junk DNA is functional. At this point, molecular geneticists have demonstrated that virtually every class of junk DNA has function. This notion undermines the best evidence for common descent and, hence, undermines an evolutionary interpretation of biology. More importantly, the discovery adds support for the competitive endogenous RNA hypothesis, which can be marshaled to support RTB’s genomics model. It is becoming more and more evident to me that genome structure and function reflect the handiwork of a Creator.
The Role of a Pseudogene in Mediating Hypoalgesia
To identify the genetic mutation responsible for the 66-year-old’s hypoalgesia, the research team scanned her DNA along with samples taken from her mother and two children. The team discovered two genetic changes: (1) mutations to the FAAH gene that reduced its expression, and (2) deletion of part of the FAAH pseudogene.
The FAAH gene encodes for a protein called fatty acid amide hydrolase (FAAH). This protein breaks down fatty acid amides. Some of these compounds interact with cannabinoid receptors. These receptors are located in the membranes of cells found in tissues throughout the body. They mediate pain sensation, among other things. When fatty acid amide concentrations become elevated in the circulatory system, it produces an analgesic effect.
Researchers found elevated fatty acid amide levels in the patient’s blood, consistent with reduced expression of the FAAH gene. It appears that both mutations are required for the complete hypoalgesia observed in the patient. The patient’s mother, daughter, and son all display only partial hypoalgesia. The mother and daughter have the same mutation in the FAAH gene but an intact FAAH pseudogene. The patient’s son is missing the FAAH pseudogene, but has a “normal” FAAH gene.
Based on the data, it looks like proper expression levels of the FAAH gene require an intact FAAH pseudogene. This is not the first time that biomedical researchers have observed the same effect. There are a number of gene-pseudogene pairs in which both must be intact and transcribed for the gene to be expressed properly. In 2011, researchers from Harvard University proposed that the competitive endogenous RNA hypothesis explains why transcribed pseudogenes are so important for gene expression.2
The Competitive Endogenous RNA Hypothesis
Biochemists and molecular biologists have long believed that the primary mechanism for regulating gene expression centered around controlling the frequency and amount of mRNA produced during transcription. For housekeeping genes, mRNA is produced continually, while for genes that specify situational proteins, it is produced as needed. Greater amounts of mRNA are produced for genes expressed at high levels and limited amounts for genes expressed at low levels.
Researchers long thought that once the mRNA was produced it would be translated into proteins, but recent discoveries indicate this is not the case. Instead, an elaborate mechanism exists that selectively degrades mRNA transcripts before they can be used to direct the protein production at the ribosome. This mechanism dictates the amount of protein produced by permitting or preventing mRNA from being translated. The selective degradation of mRNA also plays a role in gene expression, functioning in a complementary manner to the transcriptional control of gene expression.
Another class of RNA molecules, called microRNAs, mediates the selective degradation of mRNA. In the early 2000s, biochemists recognized that by binding to mRNA (in the 3′ untranslated region of the transcript), microRNAs play a crucial role in gene regulation. Through binding, microRNAs flag the mRNA for destruction by RNA-induced silencing complex (RISC).
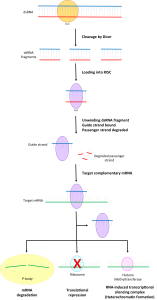
Figure 2: Schematic of the RNA-Induced Silencing Mechanism. Image Credit: Wikipedia
Various distinct microRNA species in the cell bind to specific sites in the 3′ untranslated region of mRNA transcripts. (These binding locations are called microRNA response elements.) The selective binding by the population of microRNAs explains the role that duplicated pseudogenes play in regulating gene expression.
The sequence similarity between the duplicated pseudogene and the corresponding “intact” gene means that the same microRNAs will bind to both mRNA transcripts. (It is interesting to note that most duplicated pseudogenes are transcribed.) When microRNAs bind to the transcript of the duplicated pseudogene, it allows the transcript of the “intact” gene to escape degradation. In other words, the transcript of the duplicated pseudogene is a decoy. The mRNA transcript can then be translated and, hence, the “intact” gene expressed.
It is not just “intact” and duplicated pseudogenes that harbor the same microRNA response elements. Other genes share the same set of microRNA response elements in the 3′ untranslated region of the transcripts and, consequently, will bind the same set of microRNAs. These genes form a network that, when transcribed, will influence the expression of all genes in the network. This relationship means that all the mRNA transcripts in the network can function as decoys. This recognition accounts for the functional utility of unitary pseudogenes.
One important consequence of this hypothesis is that mRNA has dual functions inside the cell. First, it encodes information needed to make proteins. Second, it helps regulate the expression of other transcripts that are part of its network.
Junk DNA and the Case for Creation
Evolutionary biologists have long maintained that identical (or nearly identical) pseudogene sequences found in corresponding locations in genomes of organisms that naturally group together (such as humans and the great apes) provide compelling evidence for shared ancestry. This interpretation was persuasive because molecular geneticists regarded pseudogenes as nonfunctional, junk DNA. Presumably, random biochemical events transformed functional DNA sequences (genes) into nonfunctional garbage.
Creationists and intelligent design proponents had little to offer by way of evidence for the intentional design of genomes. But all this changed with the discovery that virtually every class of junk DNA has function, including all three types of pseudogenes (processed, duplicated, and unitary).
If junk DNA is functional, then the sequences previously thought to show common descent could be understood as shared designs. The competitive endogenous RNA hypothesis supports this interpretation. This model provides an elegant rationale for the structural similarity between gene-pseudogene pairs and also makes sense of the widespread presence of unitary pseudogenes in genomes.
Of course, this insight also supports the RTB genomics model. And that sure feels good to me.
Resources
- Who Was Adam? A Creation Model Approach to the Origin of Humanity
- The Cell’s Design: How Chemistry Reveals the Creator’s Artistry
- “Pseudogene Decoy Reveal Hidden Evidence for Design” by Fazale Rana (article)
- “Functional Pseudogenes Are Everywhere!” by Fazale Rana (article)
- “High Levels of Pseudogene Expression Help Silence the Case for Common Descent” by Fazale Rana (article)
- “Scientific Upset: More Functions for Pseudogenes” by Fazale Rana (article)
- “Unitary Pseudogenes Have Function! Part 1” by Fazale Rana (article)
- “Unitary Pseudogenes Have Function! Part 2” by Fazale Rana (article)
- “Does the Evolutionary Paradigm Stymie Scientific Advance?” by Fazale Rana (article)
- “Evolution’s Flawed Approach to Science” by Fazale Rana (article)
Check out more from Reasons to Believe @Reasons.org
Endnotes
- Abdella M. Habib et al., “Microdeletion in a FAAH Pseudogene Identified in a Patient with High Anandamide Concentrations and Pain Insensitivity,” British Journal of Anaesthesia, advanced access publication, doi:10.1016/j.bja.2019.02.019.
- Ana C. Marques, Jennifer Tan, and Chris P. Ponting, “Wrangling for microRNAs Provokes Much Crosstalk,” Genome Biology 12, no. 11 (November 2011): 132, doi:10.1186/gb-2011-12-11-132; Leonardo Salmena et al., “A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language?”, Cell 146, no. 3 (August 5, 2011): 353–58, doi:10.1016/j.cell.2011.07.014.