Competitive Endogenous RNA Hypothesis Supports the Case for Creation
When Francis Crick, codiscoverer of the DNA double helix, first conceived of molecular biology’s organizing principle in 1958, he dubbed it the central dogma. He soon came to regret the term. In his autobiographical account, What Mad Pursuit, Crick writes:
I called this idea the central dogma, for two reasons, I suspect. I had already used the obvious word hypothesis in the sequence hypothesis, and in addition I wanted to suggest that this new assumption was more central and more powerful….As it turned out, the use of the word dogma caused almost more trouble than it was worth. Many years later Jacques Monod pointed out to me that I did not appear to understand the correct use of the word dogma, which is a belief that cannot be doubted. I did apprehend this in a vague sort of way but since I thought that all religious beliefs were without foundation, I used the word the way I myself thought about it, not as most of the world does, and simply applied it to a grand hypothesis that, however plausible, had little direct experimental support.1
Even though Crick rued labeling his idea as “dogma,” the term seems to fit, all the connotations aside, because of its singular importance to molecular biology.
The Central Dogma of Molecular Biology
The central dogma of molecular biology describes the directional flow of information in the cell, which moves from DNA to RNA to proteins. Information can flow from DNA to DNA during DNA replication, from DNA to RNA during transcription, and from RNA back to DNA during reverse transcription. However, biochemical information can’t flow from proteins to either RNA or DNA.
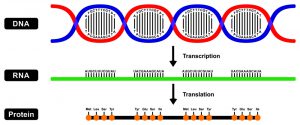
Figure 1: The Central Dogma of Molecular Biology. Image credit: Shutterstock
Is There a New Dogma in Molecular Biology?
In my opinion as a biochemist, if there is an idea that has the potential to rival the significance of the central dogma, it just might be the competitive endogenous RNA (ceRNA) hypothesis. This newer model provides a comprehensive description of the role messenger RNA (mRNA) molecules play in regulating gene expression, thereby influencing the flow of information from DNA to proteins.
The ceRNA hypothesis also provides an elegant rationale for why the genomes of eukaryotic organisms contain pseudogenes (including unitary pseudogenes) and encode long noncoding RNA molecules. Additionally, it explains why duplicated pseudogenes resemble corresponding intact genes. In doing all this, the ceRNA hypothesis provides support for the RTB’s genomics model—which interprets the structure and activities associated with genomes from a creation or design standpoint. (An overview of the RTB genomics model can be found in the updated and expanded 2nd edition of Who Was Adam?)
The Competitive Endogenous RNA Hypothesis
I discuss the ceRNA hypothesis in a previous article. So, I’ll offer just a brief description here. According to the central dogma, the final step in the flow of biochemical information is the production of proteins at the ribosome, directed by the information housed in mRNA. Biochemists have discovered an elaborate mechanism that selectively degrades mRNA transcripts before they can reach this point. This degradation process controls gene expression by dictating the amount of protein produced.
Molecules called microRNAs bind to the mRNA’s 3′ untranslated region, which flags the transcript for destruction by RNA-induced silencing complex (RISC). A number of distinct microRNA species exist in the cell. Each microRNA species bind to specific sites in the 3′ untranslated region of mRNA transcripts. (These binding locations are called microRNA response elements or MREs.)
A network of genes shares the same set of MREs and, consequently, will bind to the same set of microRNAs. When one gene is transcribed, it will influence the expression of all the other genes in its network. And when one gene in the network becomes up-regulated (leading to increased transcription of that gene), the expression of all the genes in the network increases. Why? Because the increased level of that particular transcript exerts a “sponge effect” that consumes more of the microRNAs that would otherwise target other transcripts for degradation.
The Competitive Endogenous RNA Hypothesis and the Role of Junk DNA
The ceRNA hypothesis elegantly explains the functional utility of three classes of junk DNA: duplicated and unitary pseudogenes, plus long noncoding RNAs. As it turns out, the transcripts produced from these types of so-called junk DNA also harbor MREs. None of these transcripts codes for proteins yet they play an indispensable role in regulating gene expression. In fact, all three are much better suited for the role of molecular sponges precisely because they aren’t translated into proteins.
Of particular utility are duplicated pseudogenes due to their close structural resemblance to the corresponding coding genes. Duplicated pseudogenes not only exert a sponge effect but also serve as decoys that allow the transcripts of the intact genes to escape degradation and to be translated into proteins.
Is the Competitive Endogenous RNA Hypothesis Valid?
This question has generated a minor scientific controversy. Some studies provide experimental support for this idea while others question the physiological relevance of ceRNAs. In light of this debate, a team of researchers headed by investigators from Columbia University sought to validate this hypothesis on a large-scale.2 They discovered that ceRNA interactions can disrupt the expression of thousands of genes. The team concluded that “ceRNA regulation is the norm not the exception…and that ceRNA interactions have genome-wide effects on gene expression.”3
These researchers think that this insight sheds light on tumor biology because dysregulation of ceRNAs have been implicated in some cancers. Their work also has theological significance because it undermines one of the most significant challenges to design arguments and, in turn, can be marshaled in support of the RTB genomics model.
The Competitive Endogenous Hypothesis and the Case for a Creator
Evolutionary biologists have long maintained that identical (or nearly identical) junk DNA sequences (such as pseudogene sequences) found in corresponding locations in genomes of organisms that naturally cluster together (such as humans and the great apes) provide compelling evidence that these organisms must have evolved from a shared ancestor. This interpretation was compelling because junk DNA sequences seemed to be useless vestiges of evolutionary history.
Creationists and intelligent design proponents had little to offer by way of evidence for the intentional design of genomes. But research in recent years has revealed that virtually every class of junk DNA has function. It seems, then, that shared junk DNA sequences can be understood as shared designs, which is what the RTB genomics model predicts.
Additionally, the ceRNA hypothesis supports the RTB genomics even further. This hypothesis provides an elegant explanation for the widespread existence of pseudogenes in genomes and their structural similarity to intact genes.
Could it be that the idea of religious dogma affirming a Creator’s role in life’s design and history has merit?
Resources
- Who Was Adam? A Creation Model Approach to the Origin of Humanity
- The Cell’s Design: How Chemistry Reveals the Creator’s Artistry
- “Pseudogene Decoy Reveals Hidden Evidence for Design” by Fazale Rana (article)
- “Functional Pseudogenes Are Everywhere!” by Fazale Rana (article)
- “High Levels of Pseudogene Expression Help Silence the Case for Common Descent” by Fazale Rana (article)
- “Scientific Upset: More Functions for Pseudogenes” by Fazale Rana (article)
- “Unitary Pseudogenes Have Function! Part 1” by Fazale Rana (article)
- “Unitary Pseudogenes Have Function! Part 2” by Fazale Rana (article)
- “Pseudogene Discovery Pains Evolutionary Paradigm” by Fazale Rana (article)
- “Does the Evolutionary Paradigm Stymie Scientific Advance?” by Fazale Rana (article)
- “Evolution’s Flawed Approach to Science” by Fazale Rana (article)
Check out more from Reasons to Believe @Reasons.org
Endnotes
- Francis Crick, What Mad Pursuit (New York: Basic Books, 1988), 109.
- Hua-Sheng Chiu et al., “High-Throughput Validation of ceRNA Regulatory Networks,” BMC Genomics 18 (2017): 418, doi:10.1186/s12864-017-3790-7.
- Chiu et al., 418.





