Another Disappointment for the Evolutionary Model for the Origin of Eukaryotic Cells?
We all want to be happy.
And there is no shortage of advice on what we need to do to lead happy, fulfilled lives. There are even “experts” who offer advice on what we shouldn’t do, if we want to be happy.
As a scientist, there is one thing that makes me (and most other scientists) giddy with delight: It is learning how things in nature work.
Most scientists have a burning curiosity to understand the world around them, me included. Like most scientists, I derive enormous amount of joy and satisfaction when I gain insight into the inner workings of some feature of nature. And, like most in the scientific community, I feel frustrated and disappointed when I don’t know why things are the way they are. Side by side, this combination of joy and frustration serves as one of the driving forces for my work as a scientist.
And, because many of the most interesting questions in science can appear at times to be nearly impenetrable mysteries, new discoveries typically bring me (and most other scientists) a mixture of hope and consternation.
Trying to Solve a Mystery
These mixed emotions are clearly evident in the life scientists who strive to understand the evolutionary origin of complex, eukaryotic cells. As science journalist Carl Zimmer rightly points out, the evolutionary process that produced eukaryotic cells from simpler microbes stands as “one of the deepest mysteries in biology.”1 And while researchers continue to accumulate clues about the origin of eukaryotic cells, they remain stymied when it comes to offering a robust, reliable evolutionary account of one of life’s key transitions.
The leading explanation for the evolutionary origin of eukaryotic cells is the endosymbiont hypothesis. On the surface, this idea appears to be well evidenced. But digging a little deeper into the details of this model exposes gaping holes. And each time researchers present new understanding about this presumed evolutionary transition, it exposes even more flaws with the model, turning the joy of discovery into frustration, as the latest work by a team of Japanese microbiologists attests.2
Before we unpack the work by the Japanese investigators and its implications for the endosymbiont hypothesis, a quick review of this cornerstone idea in evolutionary theory is in order. (If you are familiar with the endosymbiont hypothesis and the evidence in support of the model, please feel free to skip ahead to The Discovery of Lokiarchaeota)
According to this idea, complex cells originated when symbiotic relationships formed among single-celled microbes after free-living bacterial and/or archaeal cells were engulfed by a “host” microbe.
Much of the endosymbiont hypothesis centers around the origin of the mitochondrion. Presumably, this organelle started as an endosymbiont. Evolutionary biologists believe that once engulfed by the host cell, this microbe took up permanent residency, growing and dividing inside the host. Over time, the endosymbiont and the host became mutually interdependent, with the endosymbiont providing a metabolic benefit for the host cell, such as supplying a source of ATP. In turn, the host cell provided nutrients to the endosymbiont. Presumably, the endosymbiont gradually evolved into an organelle through a process referred to as genome reduction. This reduction resulted when genes from the endosymbiont’s genome were transferred into the genome of the host organism.
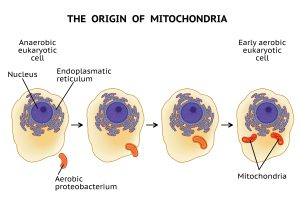
Figure 1: A Depiction of the Endosymbiont Hypothesis. Image credit: Shutterstock
Evidence for the Endosymbiont Hypothesis
At least three lines of evidence bolster the hypothesis:
- The similarity of mitochondria to bacteria. Most of the evidence for the endosymbiont hypothesis centers around the fact that mitochondria are about the same size and shape as a typical bacterium and have a double membrane structure like gram-negative cells. These organelles also divide in a way that is reminiscent of bacterial cells.
- Mitochondrial DNA. Evolutionary biologists view the presence of the diminutive mitochondrial genome as a vestige of this organelle’s evolutionary history. They see the biochemical similarities between mitochondrial and bacterial genomes as further evidence for the evolutionary origin of these organelles.
- The presence of the unique lipid, cardiolipin, in the mitochondrial inner membrane. This important lipid component of bacterial inner membranes is not found in the membranes of eukaryotic cells—except for the inner membranes of mitochondria. In fact, biochemists consider cardiolipin a signature lipid for mitochondria and another relic from its evolutionary past.
The Discovery of Lokiarchaeota
Evolutionary biologists have also developed other lines of evidence in support of the endosymbiont hypothesis. For example, biochemists have discovered that the genetic core (DNA replication and the transcription and translation of genetic information) of eukaryotic cells resembles that of the Archaea. This similarity suggests to many biologists that a microbe belonging to the archaeal domain served as the host cell that gave rise to eukaryotic cells.
Life scientists think they may have made strides toward identifying the archaeal host. In 2015, a large international team of collaborators reported the discovery of Lokiarchaeota, a new phylum belonging to the Archaea. This phylum groups with eukaryotes on the evolutionary tree. Analysis of the genomes of Lokiarchaeota reveal the presence of genes that encode for the so-called eukaryotic signature proteins (ESPs). These genes are unique to eukaryotic organisms.3
As exciting as the discovery has been for evolutionary biologists, it has also been a source of frustration. Researchers didn’t discover this group of microbes by isolating microbes and culturing them in the lab. Instead, they discovered them by recovering DNA fragments from the environment (a hydrothermal vent system in the Atlantic Ocean called Loki’s Castle, after Loki, the ancient Norse god of trickery) and assembling them into genome sequences. Through this process, they learned that Lokiarchaeota correspond to a new group of Archaea, called the Asgardians. The reconstructed Lokiarchaeota “genome” is low quality (1.4-fold coverage) and incomplete (8 percent of the genome is missing).
Mystery Solved?
So, without actual microbes to study, the best that life scientists could do was infer the cell biology of Lokiarchaeota from its genome. But this frustrating limitation recently turned into excitement as a team of Japanese microbiologists isolated and cultured the first microbe that belongs to this group of archaeons, dubbed Prometheoarchaeum syntrophicum. It took researchers nearly 12 years of laboratory work to isolate this slow-growing microbe from sediments in the Pacific Ocean and culture it in the laboratory. (It takes 14 to 25 days for the microbe to double.) But this effort is now paying off, because the research team is now able to get a glimpse into what many life scientists believe to be a representative of the host microbe that spawned the first eukaryotic cells.
P. syntrophicum is spherically shaped and about 550 nm in size. In culture, this microbe forms aggregates around an extracellular polymeric material it secretes. It also has unusual membrane-based tentacle-like protrusions (of about 80 to 100 nm in length) that extend from the cell surface.
Researchers were unable to produce a pure culture of P. syntrophicum because it forms a close association with other microbes. The team learned that P. syntrophicum lives a syntrophic lifestyle, meaning that it forms interdependent relationships with other microbes in the environment. Specifically, P. syntrophicum produces hydrogen and formate as metabolic by-products that, in turn, are scavenged for nutrients by partner microbes. Researchers also discovered that P. syntrophicum consumes amino acids externally supplied in the growth medium. Presumably, this observation means that in the ocean floor sediments, P. syntrophicum feeds on organic materials released by its microbial counterpart.
P. syntrophicum and Failed Predictions of the Endosymbiont Hypothesis
Availability of P. syntrophicum cells now allows researchers the unprecedented chance to study a microbe that they believe stands in as a representative for the archaeal host in the endosymbiont hypothesis. Has the mystery been solved? Instead of affirming the scientific predictions of leading versions of the endosymbiont hypothesis, the biology of this organism adds to the frustration and confusion surrounding the evolutionary account. Scientific analysis produces raises three questions for the evolutionary view:
- First, this microbe has no internal cellular structures. This observation stands as a failed prediction. Because Lokiarchaeota (and other members of the Asgard archaeons) have a large number of ESPs present in their genomes, some biologists speculated that the Asgardian microbes would have complex subcellular structures. Yet, this expectation has not been realized for P. syntrophicum, even though this microbe has around 80 or so ESPs in its genome.
- Second, this microbe can’t engulf other microbes. This inability also serves as a failed prediction. Prior to the cultivation of P. syntrophicum, analysis of the genomes of Lokiarchaeota identified a number of genes involved in membrane-related activities, suggesting that this microbe may well have possessed the ability to engulf other microbes. Again, this expectation wasn’t realized for P. syntrophicum. This observation is a significant blow to the endosymbiont hypothesis, which requires the host cell to have cellular processes in place to engulf other microbes.
- Third, the membranes of this microbe are comprised of typical archaeal lipids and lack the enzymatic machinery to make typical bacterial lipids. This also serves as a failed prediction. Evolutionary biologists had hoped that P. syntrophicum would provide a solution to the lipid divide (next section). It doesn’t.
What Is the Lipid Divide?
The lipid divide refers to the difference in the chemical composition of the cell membranes found in bacteria and archaea. Phospholipids comprise the cell membranes of both sorts of microbes. But that‘s where the similarity ends. The chemical makeup of the phospholipids is distinct in bacteria and archaea, respectively.
Bacterial phospholipids are built around a D-glycerol backbone which has a phosphate moiety bound to the glycerol in the sn-3 position. Two fatty acids are bound to the D-glycerol backbone at the sn-1 and sn-2 position. In water, these phospholipids assemble into bilayer structures.
Archaeal phospholipids are constructed around an L–glycerol backbone (which produces membrane lipids with different stereochemistry than bacterial phospholipids). The phosphate moiety is attached to the sn-1 position of glycerol. Two isoprene chains are bound to the sn-2 and sn-3 positions of L-glycerol via ether linkages. Some archaeal membranes are formed from phospholipid bilayers, while others are formed from phospholipid monolayers.
Presumably, the structural features of the archaeal phospholipids serve as an adaptation that renders them ideally suited to form stable membranes in the physically and chemically harsh environments in which many archaea find themselves.
The Lipid Divide Frustrates the Endosymbiont Hypothesis
If the host cell in the endosymbiont evolutionary mechanism is an archaeal cell, it logically follows that the membrane composition of eukaryotic cells should be archaeal-like. As it turns out, this expectation is not met. The cell membranes of eukaryotic cells closely resemble bacterial, not archaeal, membranes.
Can Lokiarchaeota Traverse the Lipid Divide?
Researchers had hoped that the discovery of Lokiarchaeota would shed light on the evolutionary origin of eukaryotic cell membranes. In the absence of having actual organisms to study, researchers screened the Lokiarchaeota genome for enzymes that would take part in phospholipid synthesis, with the hopes of finding clues about how this transition may have occurred.
Based on their analysis, they argued that Lokiarchaeota could produce some type of hybrid phospholipid with features of both archaeal and bacterial phospholipids. Still, their conclusion remained speculative at best. The only way to establish Lokiarchaeota membranes as transitional between those found in archaea and bacteria is to perform chemical analysis of its membranes. With the isolation and cultivation of P. syntrophicum this analysis is possible. Yet its results only serve to disappoint evolutionary biologists, because this microbe has typical archaeal lipids in its membranes and displays no evidence of being capable of making archaeal/bacterial hybrid lipids.
A New Model for the Endosymbiont Hypothesis?
Not to be dissuaded by these disappointing results, the Japanese researchers propose a new version of the endosymbiont hypothesis, consistent with P. syntrophicum biology. For this model, they envision the archaeal host entangling an oxygen-metabolizing, ATP-producing bacterium in the tentacle-like structures that emanate from its cellular surface. Over time, the entangled organism forms a mutualistic relationship with the archaeal host. Eventually, the host encapsulates the entangled microbe in an extracellular structure that forms the body of the eukaryotic cell, with the host cell forming a proto-nucleus.
Though this model is consistent with P. syntrophicum biology, it is highly speculative and lack supporting evidence. To be fair, the Japanese researchers make this very point when they state, “further evidence is required to support this conjecture.”5
This work shows how scientific advance helps validate or invalidate models. Even though many biologists view the endosymbiont hypothesis as a compelling, well-established theory, significant gaps in our understanding of the origin of eukaryotic cells persist. (For a more extensive discussion of these outages see the Resources section.) In my view as a biochemist, some of these gaps are unbridgeable chasms that motivate my skepticism about the endosymbiont hypothesis, specifically, and the evolutionary approach to explain the origin of eukaryotic cells, generally.
Of course, my skepticism leads to another question: Is it possible that the origin of eukaryotic cells reflects a Creator’s handiwork? I am happy to say that the answer is “yes.”
Resources
Challenges to the Endosymbiont Hypothesis
- “Evolutionary Paradigm Lacks Explanation for Origin of Mitochondria and Eukaryotic Cells” by Fazale Rana (article)
- “Complex Protein Biogenesis Hints at Intelligent Design” by Fazale Rana (article)
- “ATP Transport Challenges the Evolutionary Origin of Mitochondria” by Fazale Rana (article)
- “Membrane Biochemistry Challenges Route to Evolutionary Origin of Complex Cells” by Fazale Rana (article)
- “Endosymbiont Hypothesis: Things Aren’t What They Seem to Be” by Fazale Rana (article)
In Support of A Creation Model for the Origin of Eukaryotic Cells
- “Endosymbiont Hypothesis and the Ironic Case for a Creator” by Fazale Rana (article)
- “Why Do Mitochondria Have DNA?” by Fazale Rana (article)
- “Mitochondrial Genomes: Evidence for Evolution or Creation?” by Fazale Rana (article)
- “Mitochondria’s Deviant Genetic Code: Creation or Evolution?” by Fazale Rana (article)
- “Can a Creation Model Explain the Origin of Mitochondria?” by Fazale Rana (article)
- “Molecular Logic of the Electron Transport Chain Supports Creation” by Fazale Rana (article)
- “Why Mitochondria Make My List of Best Biological Designs” by Fazale Rana (article)
Check out more from Reasons to Believe @Reasons.org
Endnotes
- Carl Zimmer, “This Strange Microbe May Mark One of Life’s Great Leaps,” The New York Times (January 16, 2020), https://www.nytimes.com/2020/01/15/science/cells-eukaryotes-archaea.html.
- Hiroyuki Imachi et al., “Isolation of an Archaeon at the Prokaryote-Eukaryote Interface,” Nature 577 (January 15, 2020): 519–25, doi:10.1038/s41586-019-1916-6.
- Anja Spang et al., “Complex Archaea That Bridge the Gap between Prokaryotes and Eukaryotes,” Nature 521 (May 14, 2015): 173–79, doi:10.1038/nature14447; Katarzyna Zaremba-Niedzwiedzka et al., “Asgard Archaea Illuminate the Origin of Eukaryotic Cellular Complexity,” Nature 541 (January 19, 2017): 353–58, doi:10.1038/nature21031.
- Laura Villanueva, Stefan Shouten, and Jaap S. Sinninghe Damsté, “Phylogenomic Analysis of Lipid Biosynthetic Gene and of Archaea Shed Light on the ‘Lipid Divide,’” Environmental Microbiology 19 (January 2017): 54–69, doi:10.1111/1462-2920.13361.
- Imachi et al., “Isolation of an Archaeon.”