Just-Right RNA Structure Points to Intentional Design
He is regarded as one of the best catchers to ever play major league baseball, and yet today Yogi Berra may be more well-known for his pithy and paradoxical statements, called “Yogi-isms,” than his stellar baseball career. Berra’s famous expressions have become part of our everyday lingo, so much so that I have interacted with a few younger people who have heard some of the more well-known Yogi-isms but had no idea who Yogi Berra was.
Each Yogi-ism has a story that goes with it, offering some context for the quips. For example, when Yogi first said, “when you come to a fork in the road, take it,” he was giving his friend Joe Garagiola directions to his house in Montclair, NJ. Yogi lived on a cul-de-sac that went full circle. So, whether you went left or right at the fork you would wind up at his house.
The story behind this Yogi-ism also applies to recent insight into the physicochemical forces that influence the secondary structure of RNA molecules. This new understanding reflects the work of a team headed by physicist Ard Louis from the University of Oxford.1 As it turns out, their discovery reveals a deep-seated teleology (goal, purpose) to the structure and function of RNA molecules that evinces the intentional design of biochemical systems.
Even when this newly discovered teleology to RNA secondary structure is viewed from within the evolutionary paradigm, the case for intentional design becomes inescapable. This work indicates that RNA secondary structures do not stem from the outworking of an unguided, historically contingent evolutionary process. To put it another way, either the secondary structures of RNA molecules have been directly designed or RNA secondary structures are the inevitable outcomes of evolution. In this case, no matter what fork in the road evolution takes, it’s going to wind up at the same spot every time, indicating a purpose and directionality to the evolutionary process.
Before I describe this work and explore its philosophical and theological implications, a little background information would be helpful for some readers. For those who don’t need a brief primer on RNA structure, feel free to skip to What Determines RNA Secondary and Tertiary Structure?
Levels of RNA Structure
RNA (ribonucleic acid) plays several important roles in the cell. It serves as an intermediary between DNA and proteins, transmitting the genetic information in DNA to the ribosome, where protein synthesis takes place. RNA molecules can also carry out specific functions on their own, such as catalyzing chemical reactions. In these instances, the RNA molecules are referred to as ribozymes.
Closely related in structure to DNA, RNA is a single-stranded molecule composed of nucleotides, which consist of a nitrogen-containing base (called a nucleobase), a 5-carbon sugar (ribose), and a phosphate group. The nitrogen-containing bases in RNA are adenine (A), guanine (G), cytosine (C) and uracil (U). See figure 1.
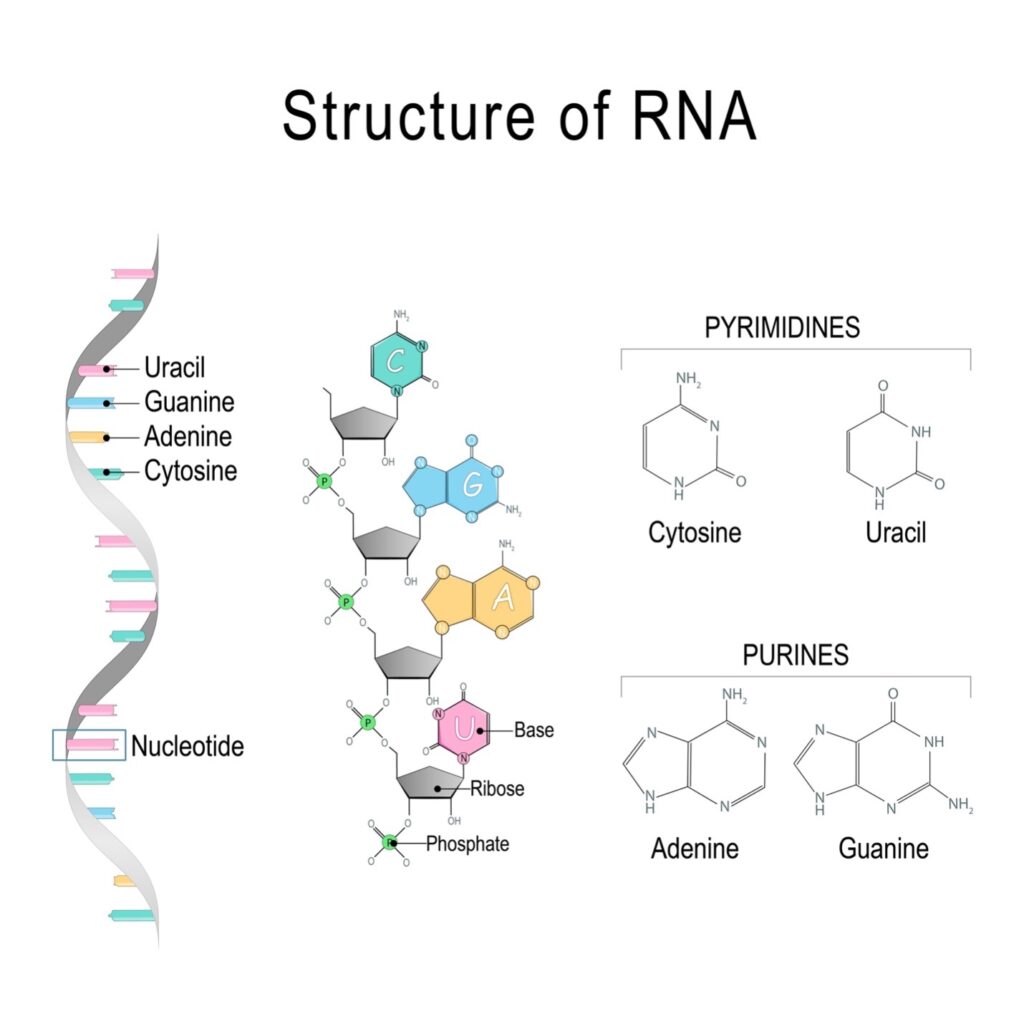
Figure 1: Structure of RNA
Credit: Shutterstock
There are three levels to RNA structure. The primary structure refers to the nucleotide sequence of the RNA strand. The secondary structure refers to the specific way the RNA strand interacts and folds upon itself. And the tertiary structure refers to the specific three-dimensional location in space of each atom of the RNA molecule.
The three levels are nicely illustrated by transfer RNA (tRNA). This biomolecule plays a critical role in protein synthesis. It is relatively small, typically around 75–90 nucleotides in length (primary structure). The intrachain interactions create a characteristic clover leaf shape (secondary structure), which is composed of several structural elements: the acceptor stem, the anticodon loop, the D-loop, and the TΨC loop. See figure 2.
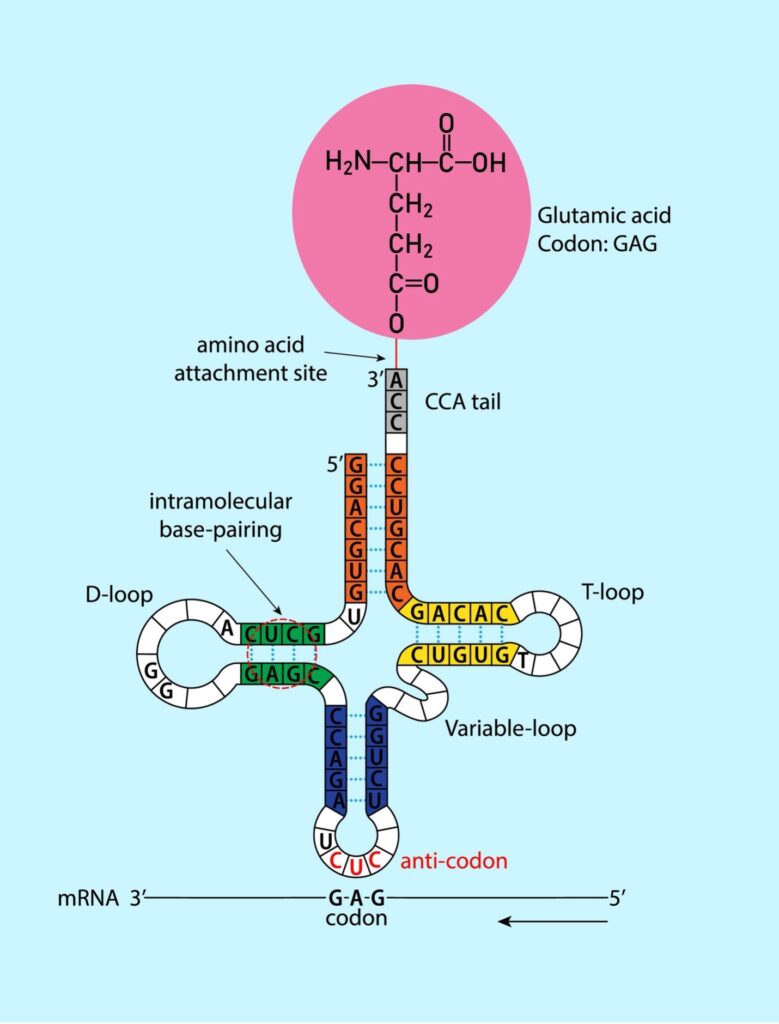
Figure 2: Secondary Structure of RNA
Credit: Shutterstock
The acceptor stem binds a specific amino acid. The anticodon loop contains the anticodon, a three-nucleotide sequence that base-pairs with a codon on the mRNA. The D-loop is a short stretch of RNA that is thought to stabilize the tRNA structure, while the TΨC loop is involved in attaching the tRNA, charged with its amino acid, to the ribosome.
The tRNA clover leaf folds into an L-shaped three-dimensional configuration. This specific shape allows the charged tRNA molecule to function as an adaptor molecule, carrying specific amino acids to the ribosome and aligning them with the codons on the mRNA so they can be assembled into a protein chain. See figure 3.
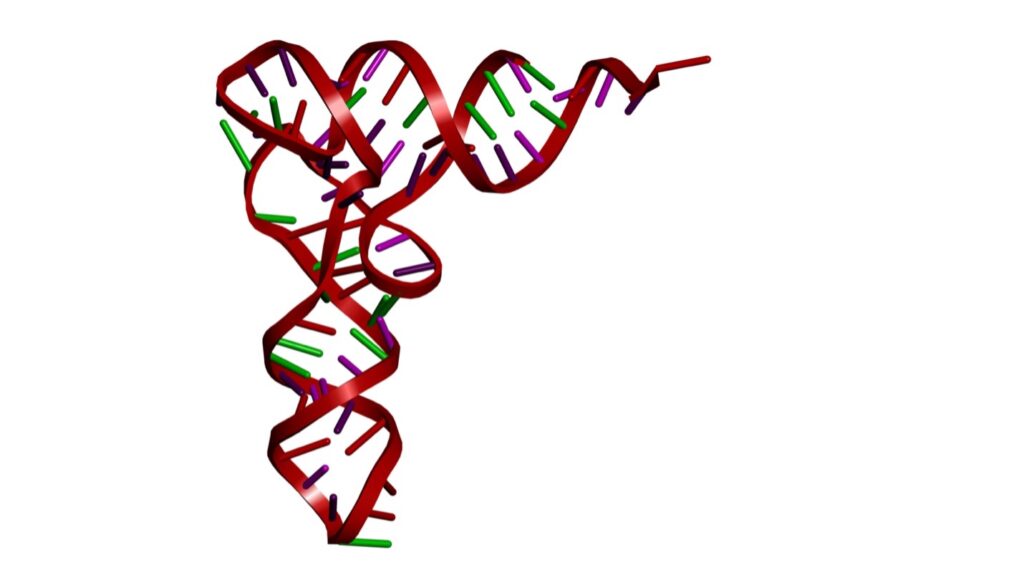
Figure 3: Tertiary Structure of RNA
Credit: Shutterstock
What Determines RNA Secondary and Tertiary Structure?
RNA molecules found inside the cell adopt a wide range of secondary and tertiary structures. The research team hoped to gain insight that will help explain the basis for this varied RNA structure.
One possibility: RNA structure can be accounted for by natural selection operating on the random variation of the RNA primary structures, in which the variation produces a vast number of secondary and tertiary structures, all with equal likelihood of occurring. An alternative possibility: RNA secondary and tertiary structures are determined by physicochemical forces, in which only certain secondary and tertiary structures are possible.
In the first scenario, natural selection dictates higher-order RNA structure. In the second scenario, the secondary and tertiary structures are constrained by the laws of nature. To simplify their work, Louis’s team focused on RNA secondary structure (which is a determinant of RNA tertiary structure). They first interrogated the RNAcentral database, cataloging all known RNA secondary structures found in nature.
They then compared the RNA secondary structures in nature with those generated from RNA molecules consisting of random nucleotide sequences for RNA chain lengths of 40, 55, 70, 85, 100, and 126 nucleotides, respectively. The researchers classified the folds into a set of abstract shapes.
They did the same procedure for the RNA molecules contained in the RNAcentral database and discovered that the RNA molecules found in nature account for a small subset of all the possible shapes that principally could exist. The team estimated that somewhere between 103 to 106 nucleotide sequences account for all the RNA shapes found in nature. For context, there are 1075 possible sequences for an RNA molecule 126 nucleotides in length.
The RNA shapes they recovered from the set of random RNA sequences were the same as those that occur for the RNA molecules found in nature. They also discovered that the frequency of the shapes generated from RNA molecules composed of random nucleotide sequences was the same frequency as the shapes of naturally occurring RNA molecules.
In other words, the researchers discovered that a bias exists regarding RNA structure. Without the influence of natural selection, the shapes found in RNA molecules comprised of random nucleotide sequences are the same as those found in the RNAcentral database.
That is to say, the shapes of RNA molecules have been presculpted. The shapes of naturally occurring RNA molecules are not the outworking of an unguided historically contingent evolutionary process. Nor have they been shaped by natural selection. The limited number of RNA shapes that occur in nature reflect physicochemical constraints.
From an evolutionary perspective, if natural selection plays any type of role, it is to fine-tune nucleotide sequences of RNA for a specific function, but it operates on shapes that have been prescribed by the laws of nature as innately existing forms.
Origin and Design of Hammerhead Ribozyme
Louis’s team’s discovery finds corroboration in an earlier study published in 2001 by Nobel Laureate Jack Szostak and a colleague.2 These two investigators were trying to understand the origin and design (from within the evolutionary paradigm) of an RNA molecule called the hammerhead ribozyme. (This biomolecule catalyzes reversible RNA cleavage and RNA ligation reactions.) The shape of this ribozyme molecule—the motif—occurs widely in organisms across all domains and kingdoms of life. Based on its distribution among organisms, it appears as if the hammerhead ribozyme arose independently, multiple times.
Using in vitro selection experiments, the two researchers screened RNA molecules with random, unbiased nucleotide sequences with the capacity for self-cleavage. They discovered that those RNA molecules with the capacity for self-cleavage had the hammerhead ribozyme motif, even though their nucleotide sequence varied. They conclude: “Our results suggest that the evolutionary process may have been channelled, in nature as in the laboratory, towards repeated selection of the simplest solution to a biochemical problem. . . . purely chemical constraints (that is, the ability of only certain sequences to carry out particular functions) can lead to the repeated evolution of the same macromolecular structures.”3
RNA Molecules: Fit for a Purpose
This understanding of RNA secondary structure developed by Ard Louis and his collaborators adds support for the biochemical anthropic principle and, with it, the intentional design of biochemical systems. In my book Fit for a PurposeI make the case that, just like the constants of physics, the core biochemical systems are finely tuned. One implication of this fine-tuning is that biochemical systems—the foundational systems of life—are the product of a Mind.
In my book, I present three criteria that must be satisfied for the anthropic principle to extend to biochemical systems:
- The molecular systems that comprise biochemical systems have been predetermined by the laws of physics and chemistry. These systems exist as natural forms. They aren’t shaped by natural selection. They aren’t the outworking of a historically contingent evolutionary history.
- The properties of these systems are precisely those that are needed for life.
- There are no legitimate alternates to these systems.
The work by Ard Louis and his team satisfies the first criterion. The RNA secondary structures that appear in nature are natural forms that are prescribed by the laws of nature.
The research team also demonstrated that a limited number of RNA shapes are found in nature (between 103 to 106). Yet, these shapes are obviously sufficient to satisfy the biochemical needs of the cell. That is to say, the natural forms of the RNAs that occur in nature are precisely those needed for life (second criterion).
Louis’s research team’s study doesn’t have any bearing one way or the other on the third criterion. However, in Fit for a Purpose, I make the case that no viable alternatives exist for RNA molecules, thereby satisfying the third criterion.
It is fortuitous—and a bit suspicious—that laws of nature have dictated the shapes of RNA molecules to be the very ones required for life. It isn’t unreasonable to think that this coincidence is more than luck. In fact, it points to a Mind that intentionally designed the universe to harbor life.
Structuralism and Creation
This discovery also provides support for a view called structuralism. On this view, evolutionary outcomes aren’t shaped by an unguided, undirected historically contingent process shaped by natural selection. Instead, evolutionary outcomes are constrained and fundamentally prescribed by the laws of nature.
According to structuralism, physical forces supersede natural selection altogether, shaping the development of biochemical and biological systems. Some structuralists go so far as to claim that the basic forms of the natural world—“the Types”—are immanent in nature and determined by a set of special natural biological laws, the so-called “laws of form.”
This view of the evolutionary process reintroduces teleology into biology. When viewed as a historically contingent process, evolution is unguided and undirected. It has no purpose. It has no end goal. On the other hand, if structuralism is valid—as the evidence increasingly suggests—then the evolutionary process proceeds to predetermined ends (such as the just-right shapes of RNA molecules) and a teleology perfuses life’s evolutionary history.
One interpretation of structuralism is that a divine Mind rigged the universe in such a way that biochemical and biological systems are inevitable and intended by the Creator. They are also the only ones that could ever emerge from evolutionary processes (like Yogi’s fork in the road).
While I don’t necessarily subscribe to structuralism, I do find the idea intriguing. Structuralism forces a singular conclusion: a Mind is behind it all—whether one adopts a creation model/design approach or an evolutionary approach to biology, the necessary role a Creator has played in life’s history is unavoidable.
With insights such as this, evolution’s fork in the road leads to the same destination in the teleological cul de sac.
Resources
- Fit for a Purpose: Does the Anthropic Principle Include Biochemistry? by Fazale Rana (book)
Biochemical Anthropic Principle
- “DNA’s Fine-Tuned Structure Minimizes Harmful Tautomers” by Fazale Rana (article)
- “The Logic of DNA Replication Makes a Case for Intelligent Design” by Fazale Rana (article)
- “How the Central Dogma of Molecular Biology Points to Design” by Fazale Rana (article)
- “Is the Optimal Set of Protein Amino Acids Purposed by a Mind?” by Fazale Rana (article)
- “A Periodic Table for Protein Structures Reveals Biochemical Design” by Fazale Rana (article)
- “Evidence That the Cell’s Metabolism Is Planned” by Fazale Rana (article)
- “Krebs Cycle Origin Brings Case for Creation Full Circle” by Fazale Rana (article)
- “Molecular Logic of the Electron Transport Chain Supports Creation” by Fazale Rana (article)
- “New Mechanism Moves Biochemical Cargo and Points to Design” by Fazale Rana (article)
Structuralism
- “What If . . . ? Convergence Strengthens the Case for Creation?” by Fazale Rana (article)
Check out more from Reasons to Believe @Reasons.org
Endnotes
1. Kamaludin Dingle et al., “Phenotype Bias Determines How Natural RNA Structures Occupy the Morphospace of All Possible Shapes,” Molecular Biology and Evolution 39, no. 1 (September 20, 2021): msab280, doi:10.1093/molbev/msab280.
2. Kourosh Salehi-Ashtiani and Jack W. Szostak, “ In vitro Evolution Suggests Multiple Origins for the Hammerhead Ribozyme,” Nature 414 (November 1, 2001): 82–84, doi:10.1038/35102081.
3. Salehi-Ashtiani and Szostak, “In vitro Evolution.”





